Fungi, plants, and microorganisms produce an enormous variety of chemical compounds that are not essential for their ground functions â the secondary metabolites. For some, a function as antibiotics, defence compounds, or pheromones, but for their majority, the function has remained enigmatic. To develop a deeper understanding of secondary metabolism, it is necessary to address the underyling metabolic pathways. In the last century, isotope labelling experiments demonstrated that secondary metabolites are composed from a limited number of building blocks (often acetate, amino acids, isoprene, or shikimate) that are subsequently modified in numerous ways. Thus, Nature plays a kind of âchemical LEOGâ by combining a limited repertory of building blocks in a modular manner to generate great chemical diversity. Biosynthetic processes that generate diversity, are, therefore, of prominent importance. Research in the lab of ALUF-MedPharm (University of Freiburg, Pharmaceutical Institute, Prof. Dr. Michael MĂŒller) ventures to elucidate biosynthetic processes that are difficult to achieve by mere chemistry. A special focus is laid on the generation of metabolic diversity and, thus, the biocatalytic exploration of entire compound classes.
This is exemplified by the enzyme-driven oxidative coupling of phenolic moieties, where identical monomers can give rise to different dimers (Fig. 1). Still at the turn of the millenium, the underlying enzymes were mostly unknown. Especially through genomic analysis, we and other groups succeeded to identify a couple of phenol-coupling enzymes and to partially describe them.
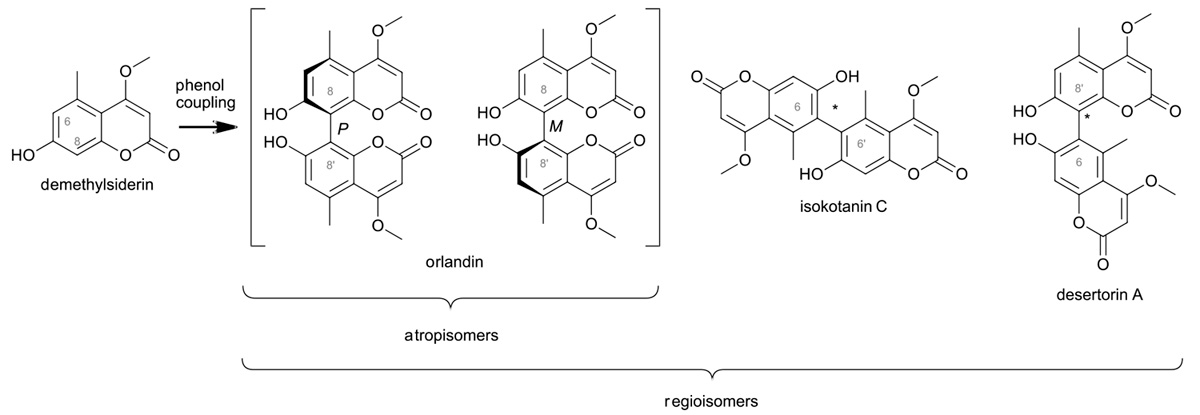
Fig. 1: enzyme-driven oxidative coupling of phenolic moieties
This showed that phenolic coupling can be catalysed by different oxidation systems, especially cytochrome-P450-enzymes and laccases, but also by peroxidases and possibly by flavin-dependent oxidases. While some of these enzymes are strictly substrate specific, others (that can be even closely related) indulge in promiscuity and accept several substrates. Which reaction is preferred, can depend from additional proteins, but also from the reaction conditions.
On this background, we will try in frame of this project to identify from bioactive fungal extracts the responsible molecules by means of activity-guided fractionation and, subsequently, to elucidate the structures of the responsible compounds. Of special interest are here the NADPH-dependent Oxidoreductases that account for the synthesis of polyketides, a central group of fungal secondary metabolites. In the next step, either these compounds, or variants of them generated by the aforementioned «chemical LEGO» via chemical synthesis or semi-synthesis, will be generated to get material for functional tests. In addition, there is the option, to link this molecular modularity with the modularity of the chip system, i.e. to simulate chemical reaction of interest on a microfluidic platform.
Reference
L. FĂŒrtges, S. Obermaier, W. Thiele, S. Foegen and M. MĂŒller, Diversity in Fungal Intermolecular Phenol Coupling of Polyketides â Regioselective Laccaseâbased systems. ChemBioChem, 2019, 20, 1928â1932. doi.org/10.1002/cbic.201900041.
S. Obermaier and M. MĂŒller, Biaryl-Forming Enzymes from Aspergilli Exhibit Substrate-Dependent Stereoselectivity, Biochemistry, 2019, 58, 2589â2593. doi.org/10.1002/chem.201705998
S. Obermaier, W. Thiele, L. FĂŒrtges and M. MĂŒller, Enantioselective Phenol Coupling by Laccases in the Biosynthesis of Fungal Dimeric Naphthopyrones. Angew. Chem. 2019, 58, 9125â9128. doi.org/10.1002/anie.201903759.
